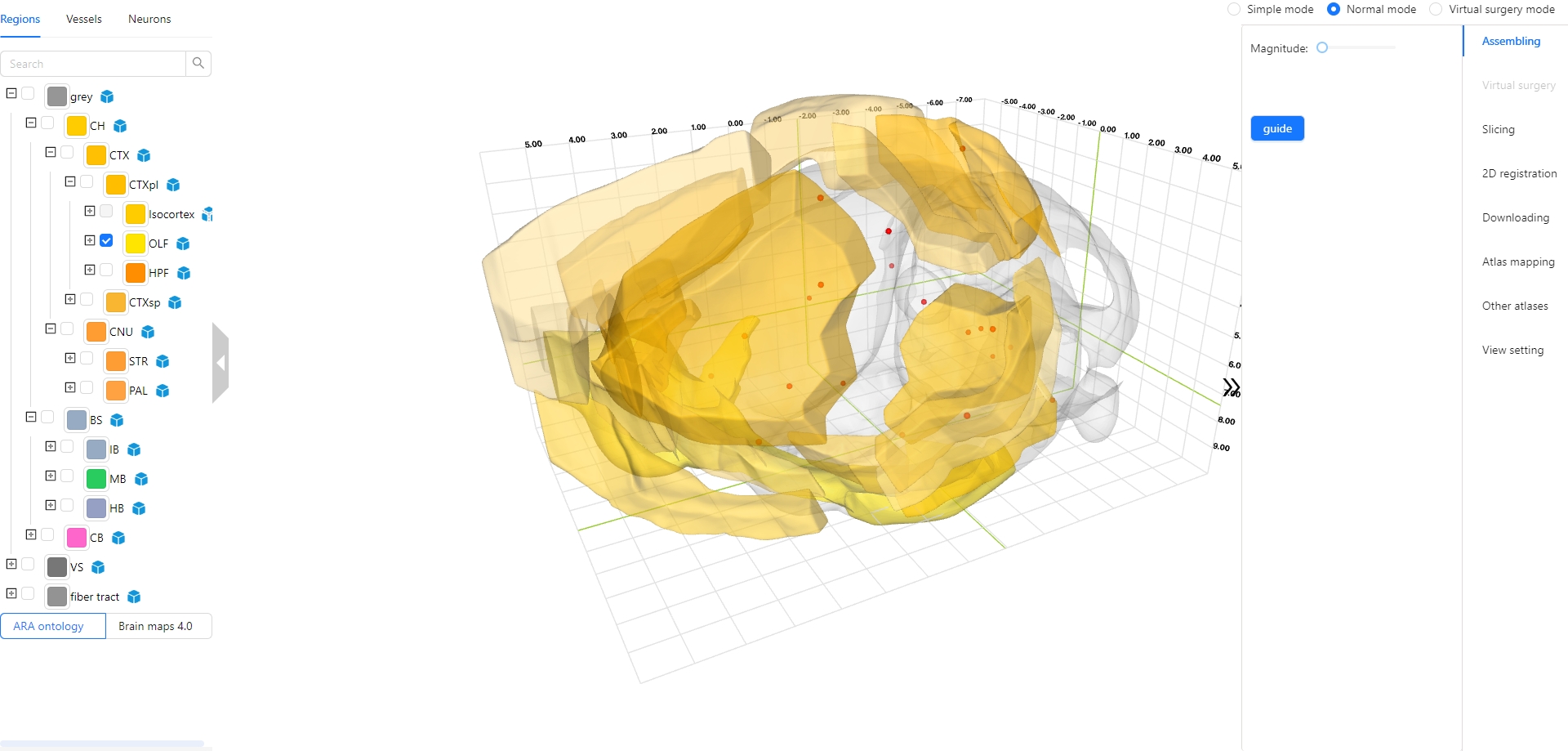
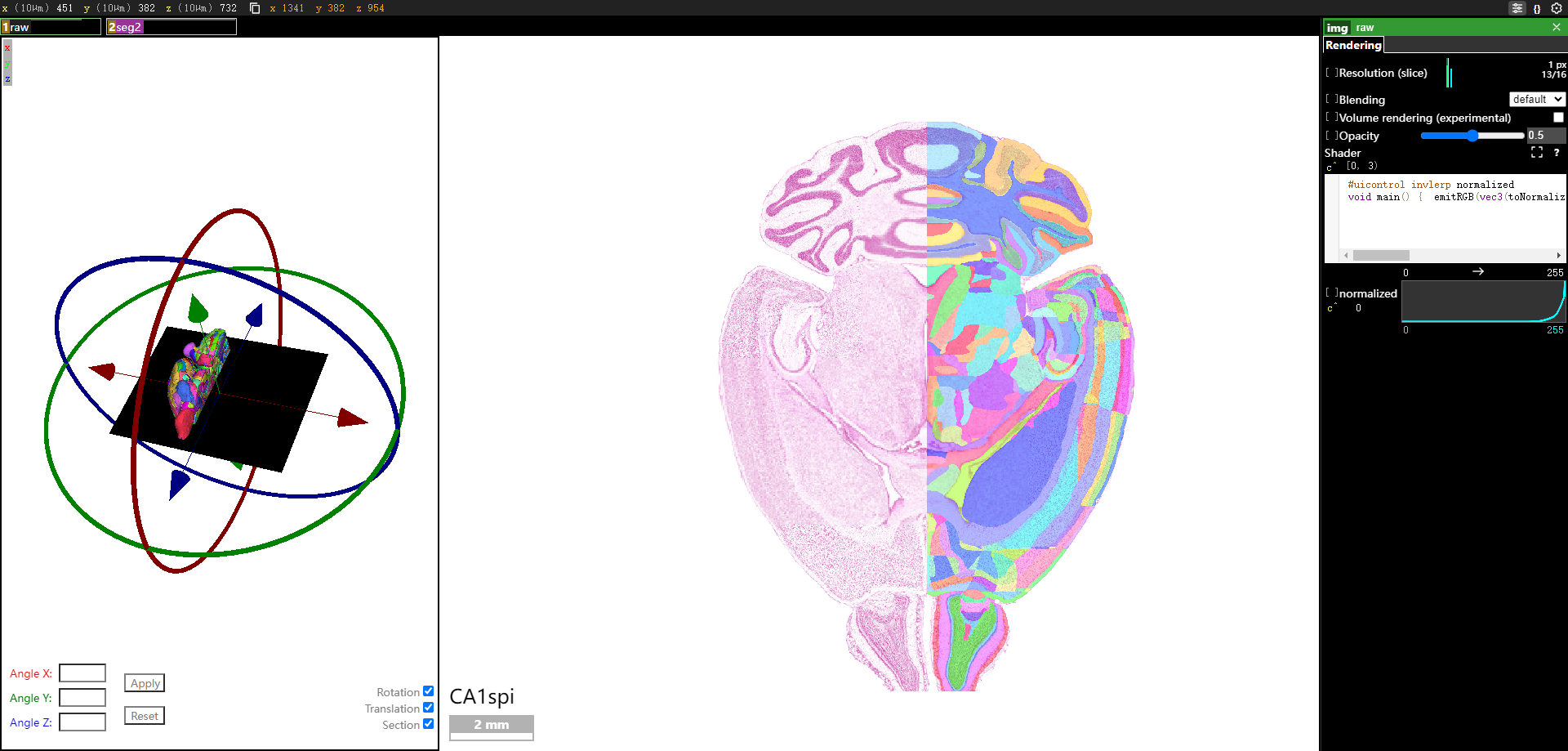
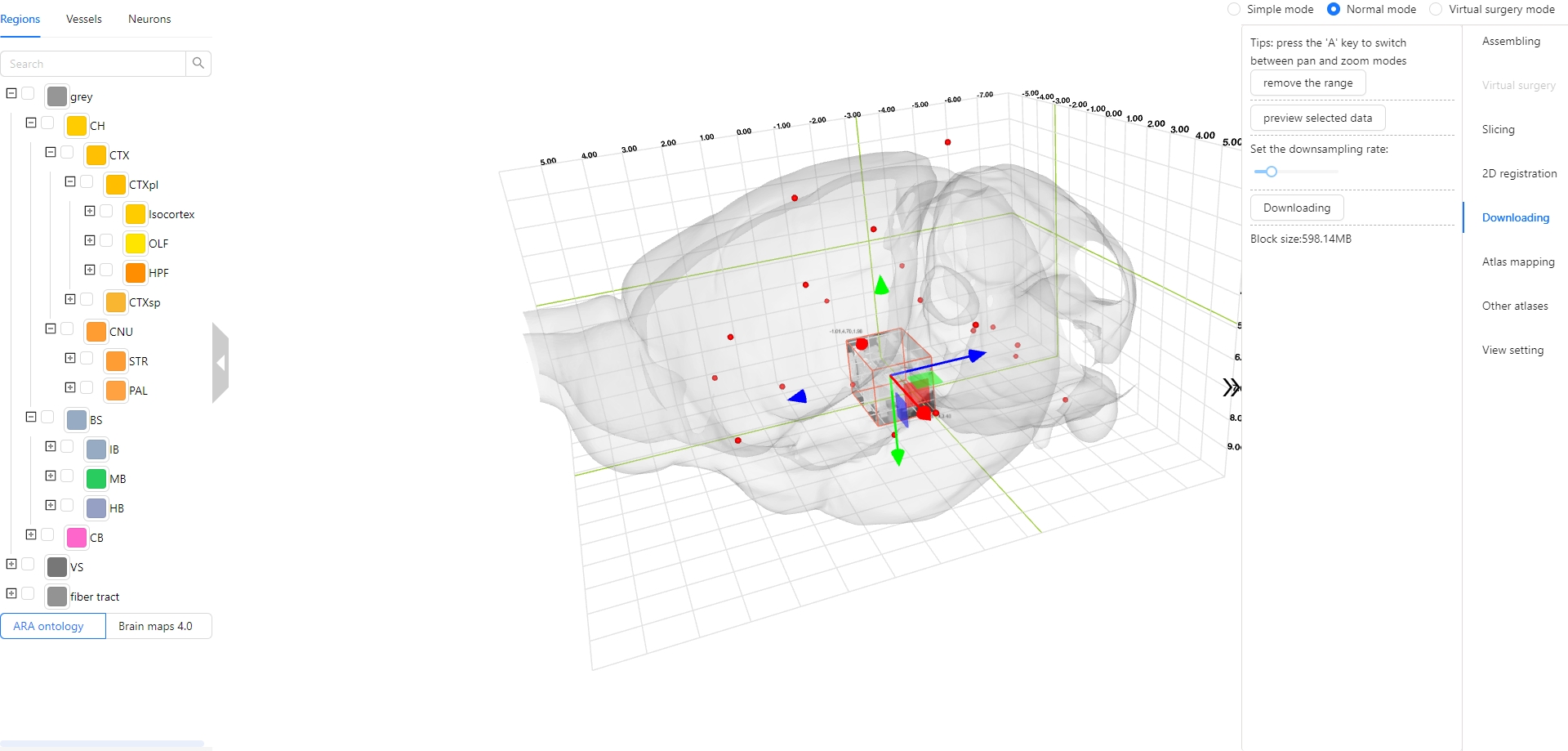
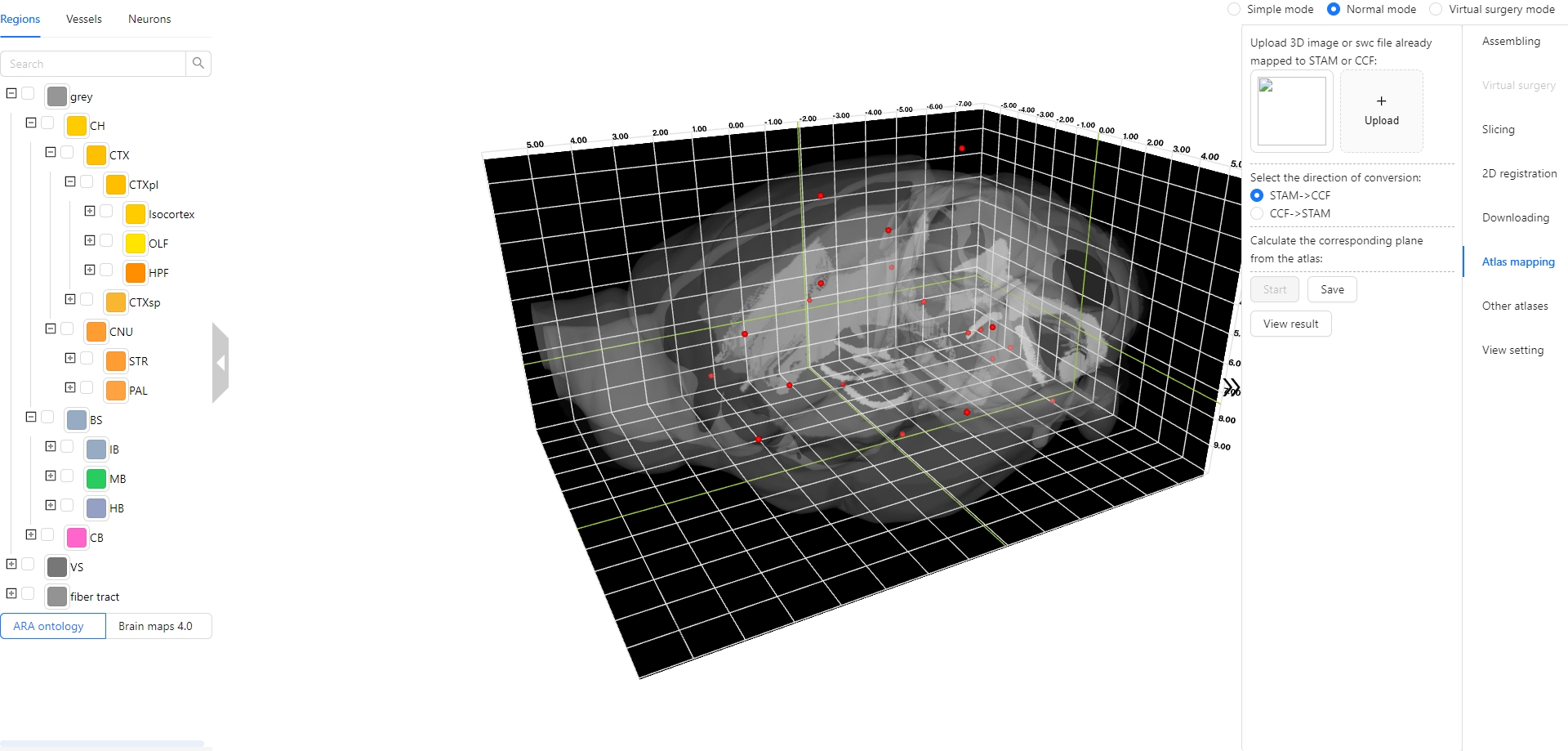
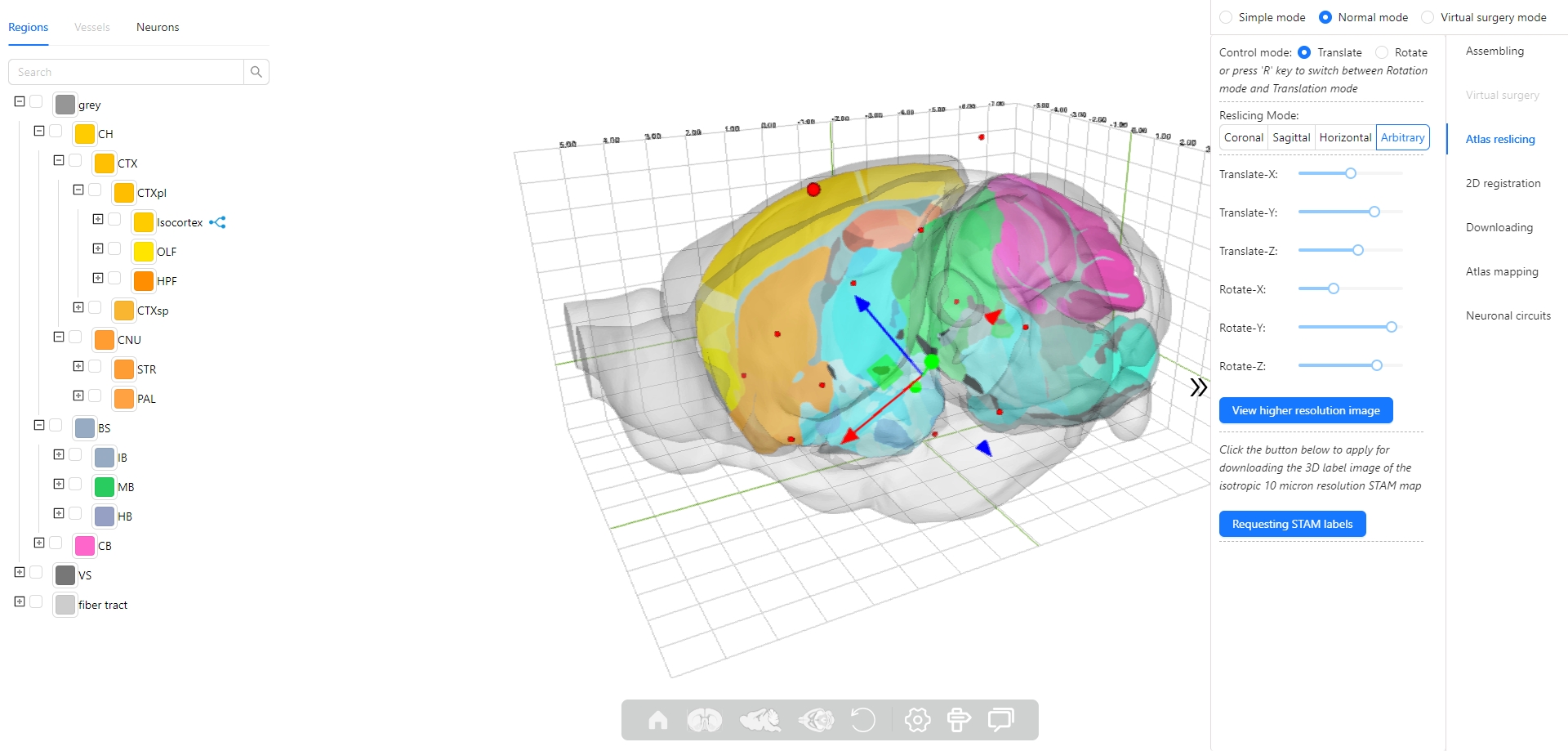
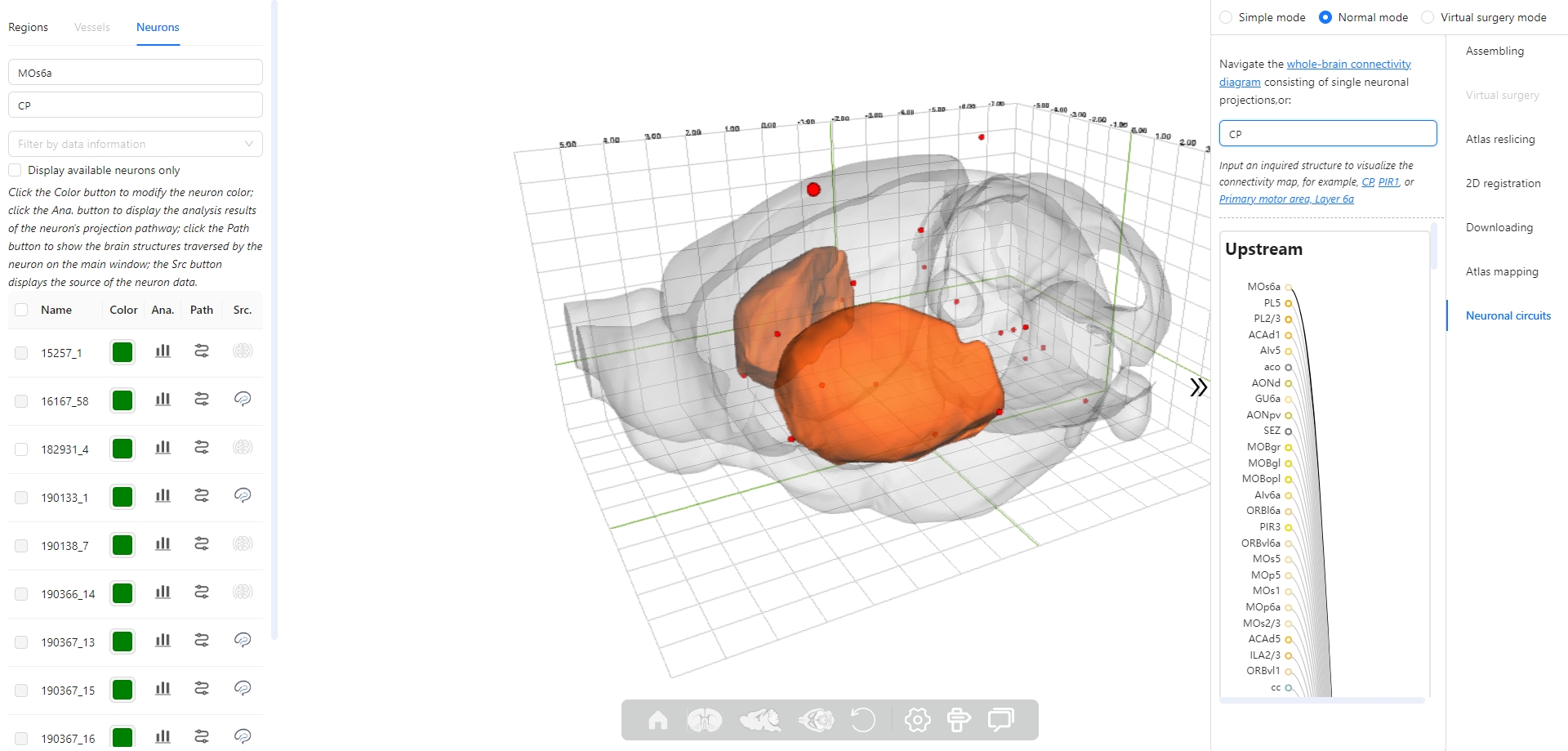
The Stereotaxic Topographic Atlas of the Mouse Brain (STAM) is a three-dimensional topographical atlas that depict the anatomical structures of the whole mouse brain, achieving precise single-cell resolution localization. We employed a three-dimensional Nissl-stained image dataset with isotropic one-micron resolution as the basis, and created canonical anatomical sections through a projection with a thickness of 20 microns resolution for constructing the STAM that contains 916 brain structures, with reference to various auxiliary imaging data, including immunohistochemistry and specific gene type neuron distribution data. Here, we established the STAM visualization platform, which includes services for exploring two-dimensional canonical anatomical sections, three-dimensional topography, and arbitrary-angle slices of the brain atlas. The platform integrates different functionalities such as virtual surgery, and automatic registration of user-uploaded slices. The STAM is expected to provide an effective spatial localization benchmark for the registration and integration of results from single-cell level studies, such as multi-omics and neuronal circuitry.
1. How to adjust the appearance of the atlas
2. How to better observe objects of interest in 3D space by setting a focus?
3. How to save the current 3D scene
4. Can I change the color of visualized objects?
5. How to interact with higher-order brain regions
6. How to interact with neurons in the 3D scene?
7. How to view quantitative analysis results of neurons?
8. How to use the atlas reslicing module?
2025.08.14
1. Canonical plane visualization : Download option for the ARA ontology tree structure is available.
2. Automatic slice registration : Support the registration for 24-bit/RGB PI-stained images.
3. Neuronal circuits : CSV format generation supported.
4. Atlas mapping : An option of determining the vertical dimension of CCF has been added.
5. Atlas mapping : The selection of spatial resolution from 1 µm to 100 µm is supported when uploading an SWC file.
2025.08.28
The Virtual surgery mode has been updated as followed:
1. The XYZ axes are labeled in the 3D scence.
2. The texts "Target" and "Injection" are labeled along the selected target site and injection site respectively.
3. The name of the selected datum mark has been added in the control panel on the right.
4. A Path-constrint mode has been provided, in which the planned path will be constraint within a selected coronal or sagittal plane.